|
|
|
|
|
We use the model plant Arabidopsis thaliana (Mouse Ear Cress, Ackerschmalwand) to investigate the subcellular localisation, molecular organisation and physiological functions of amino acid metabolism.
Our second focus area is mRNA processing as a means to regulate gene expression. We investigate both endogenous mechanisms and artifical systems of regulated mRNA processing.
Roles and Regulation of Amino Acid Metabolism in Plants
Currently, our research focuses on three aspects:
a) How does accumulation of proline contribute to the defence against drought, salt and cold stress?
b) In which ways do proline and arginine metabolism interfere and what is the physiological function of arginine catabolism?
c) How are proline and arginine transported across intracellular membranes?
Regulated mRNA processing
d) What is the physiological function of mRNA 3'-UTR processing regulation by the RSR1/ESP1 protein?
e) We are developing and characterising artificial riboswitches as tools to regulate gene expression.
Regulation and regulatory functions of proline metabolism
Drought and salinisation impose an increasing thread on agriculture and food production in many parts of the world. It is therefore an important task to understand how plants defend themselves against damage by water limitation or high osmolarity. Many plant species accumulate proline or other compatible osmolytes in response to drought or salinity stress. The exact mechanisms of how these substances protect the cells are not yet fully understood. Suggestions range from increasing the osmotic potential and stabilising membranes and proteins to the scavenging of reactive oxygen species and providing a sink for surplus chemical energy.
We use a combination of molecular genetics and physiological approaches to identify the role of proline metabolism in stress resistance. Transgenic plants with reduced or increased activity of enzymes involved in proline synthesis and degradation are compared to wildtype plants with respect to their stress responses and tolerance. With the help of protein interaction studies and mutagenesis screens we aim to identify signalling cascades that are triggered by proline or intermediates of proline metabolism.
|
|
|
 |
|
|
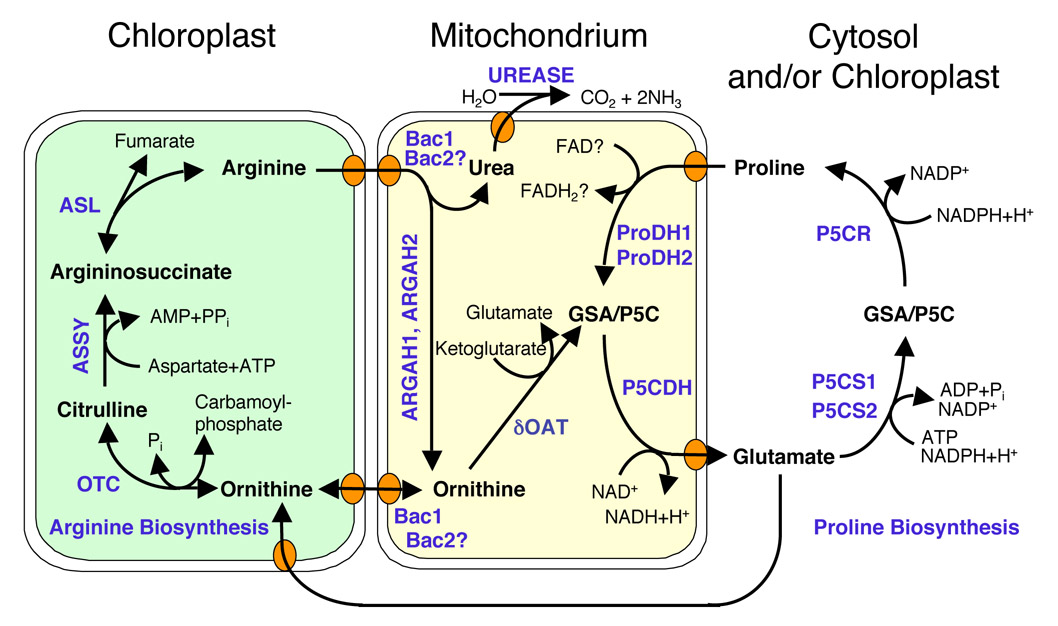
Subcellular compartmentation of proline and arginine metabolism in plants.
Most likely, the biosynthetic routes separate after glutamate, while the catabolic pathways converge in a common intermediate, pyrroline-5-carboxylate, inside mitochondria. Transporters mediating arginine and ornithine transport across the mitochondrial membrane were identified, whereas plastidic transporters and intracellular proline transporters are still unknown. |
|
|
Interactions between proline and arginine catabolism
Arginine catabolism has often been proposed to contribute to proline biosynthesis, a hypothesis that is rather unlikely when the subcellular compartmentation of both pathways is considered. We use protein interaction studies and biochemical assays to investigate the degree of overlap between proline and arginine degradation pathways. With the help of mutants in arginine degrading enzymes, we will define the physiological relevance of this pathway as well as potential alternative pathways.
Back to the top
Subcellular distribution of proline and arginine
Separation of biosynthesis and degradation of proline and arginine to different subcellular compartments generates the necessity for membrane transport of both amino acids. The need for transport is emphasized by the fact that translation and protein degradation occur independently in the cytosol, in plastids and in mitochondria. We are using candidate gene approaches and functional cloning in heterologous systems to identify transporters for proline and arginine in intracellular membranes. With the help of GFP-fusion-constructs and fluorescence microscopy we determine the subcellular localization of the respective transport proteins.
Back to the top
The role of RSR1/ESP1 in regulated mRNA 3'-end processing
The RSR1/ESP1 protein is a plant specific homolog of the mRNA Cleavage Stimulatory Factor subunit 64 (CstF64). We characterised rsr1 mutants and analyse the molecular interactions of RSR1/ESP1 with other components of the 3'-end processing aparatus
|
|
|
 |
|
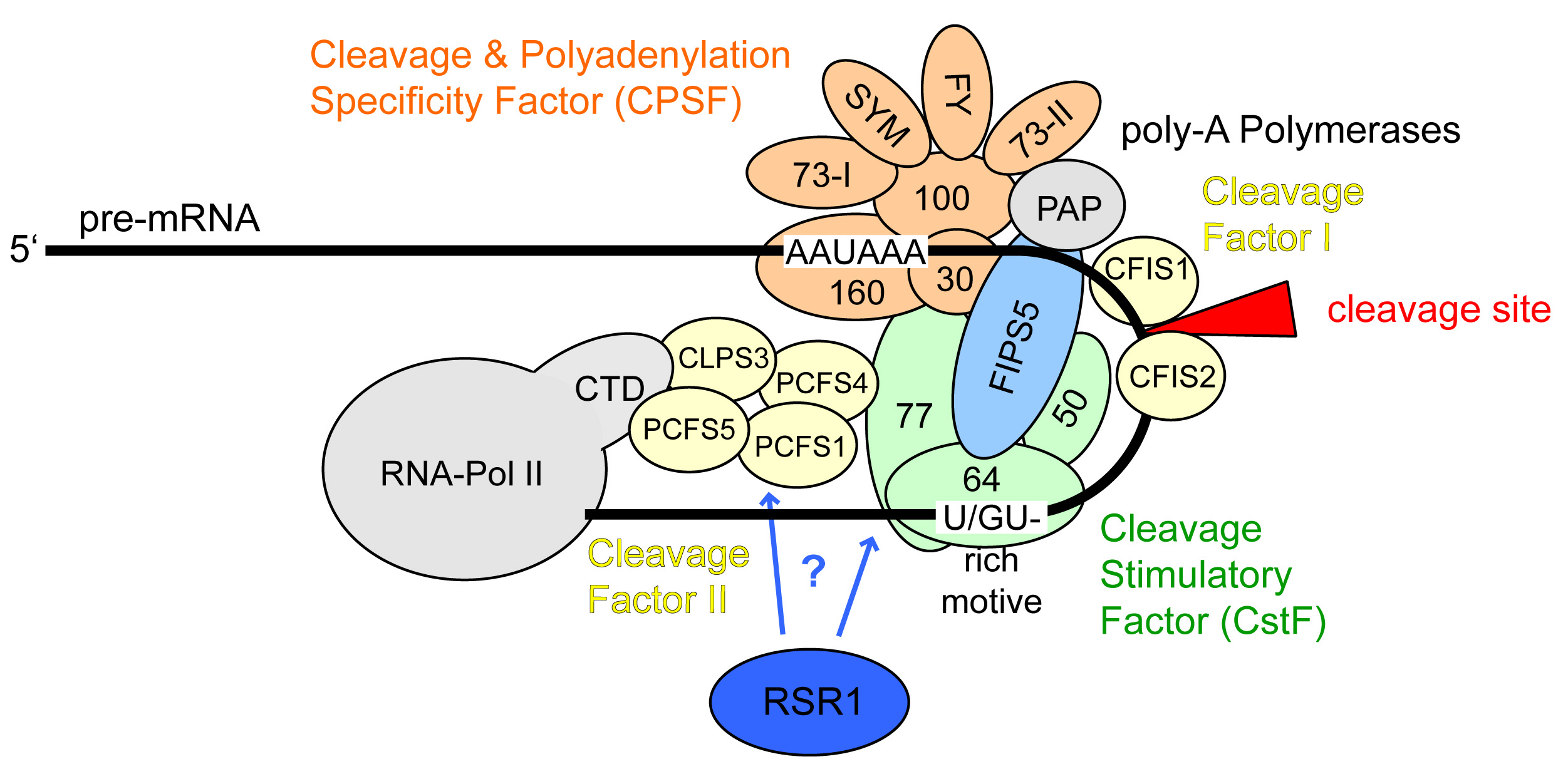
Protein complexes regulating mRNA 3'-end processing.
In a coordinated manner, transcript cleavage, polyadenylation and transcription termination are regulated by a dynamic array of multi-subunit protein complexes. |
|
|
Back to the top
Development of artificial riboswitches for regulation of gene expression
In collaboration with the AG Hartig from the Chemistry department, we are developing and characterising artificial riboswitches as tool to regulate transgene expression in plants. Riboswitches typically act in cis and very few components are needed to achiev efficient switching of gene expression. We use natural and synthetic ligand-binding RNA aptamers in combination with self-cleaving rybozymes at different positions within mRNAs to achieve stringent control of mRNA stability. |
|
|
 |
|
|
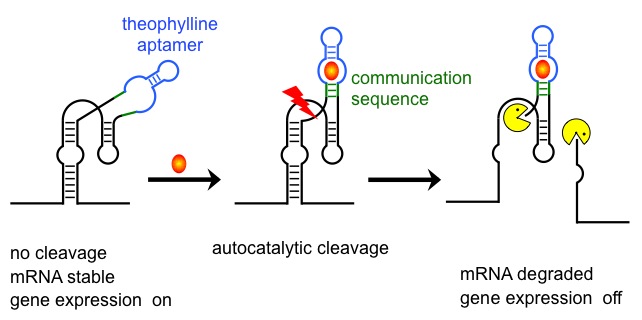
Artificial riboswitch in action.
Bnding of a specific ligand to an RNA aptamer seqence induces a conformatioal change that activates or inctivates autocatalytic cleavage in the core of a ribozyme. Cleavage produces free mRNA 3'-termini and de-stabilises the transcript. |
|
| Back to the top |
|
|
|
|
|
|
|
|
|