The untapped nitrogen reservoir
Konstanz-based research team elucidates how bacteria use the compound guanidine as a source of nitrogen
Guanidine is one of the most nitrogen-rich compounds. It could be a valuable source of organic nitrogen, but only very few organisms can access it. However, certain bacteria manage to obtain nitrogen from guanidine. A Konstanz-based research team led by chemist Professor Jörg Hartig and biologist Professor Olga Mayans has now discovered how this works. A newly discovered enzyme plays a key role – and, surprisingly, so does nickel. The research results were published on 9 March 2022 in the scientific journal Nature.
No growth without nitrogen
Nitrogen is an important component of all living organisms, and no growth is possible without nitrogen uptake. Although almost 80 percent of the atmosphere are nitrogen, the vast majority of life forms cannot access this reserve. They are thus dependent on chemically bound nitrogen, which is therefore also a pivotal component of fertilisers. However, where there is not enough nitrogen available, plants as well as many microorganisms quickly reach their limits.
There are nitrogen reserves in nature that are barely utilized: Guanidine is a widespread nitrogen-rich compound that excels by particularly high chemical stability. Due to this stability, it is hardly possible for organisms to obtain the vital nitrogen from guanidine: They cannot “crack the nut”, so to speak. Hence, many organisms are within reach of an abundant source of nitrogen – and yet cannot tap it.
A Konstanz-based research network led by chemist Professor Jörg Hartig and biologist Professor Olga Mayans has now identified a biochemical mechanism that enables certain microorganisms to extract nitrogen from guanidine. In nitrate-poor environments, this is a decisive advantage over competing organisms.
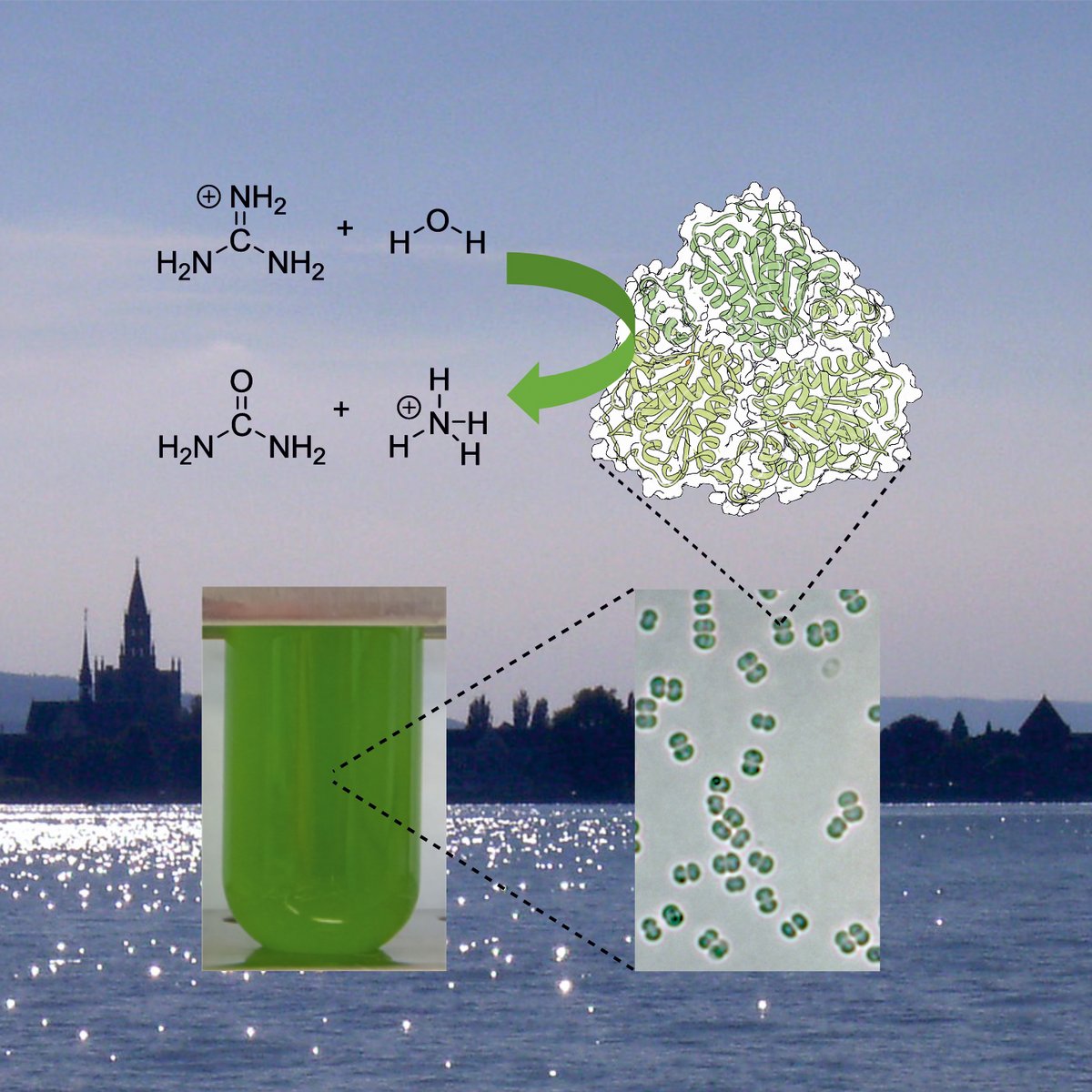
How the nitrogen mining works
Cyanobacteria, also known as blue-green algae, use an enzyme from the arginase family to initiate degradation of guanidine in the form of hydrolysis. Hydrolysis initially means merely the splitting of a chemical compound by water. In the case of guanidine, however, contact with water alone is not enough: "When guanidine is immersed in water, for hundreds of millennia virtually nothing will happen – because there is not enough energy to attack this compound," says Dr Dietmar Funck, a biologist from Konstanz.
The water therefore first needs to be "primed" in order to become chemically more active and be able to break down the guanidine. This is done by binding to nickel ions. The fact that, of all things, nickel is used as the catalyst came as a surprise to the research team. "Nickel is special. Nickel is complicated. Very quickly you have either too little or too much of it," describes Jörg Hartig: "We humans no longer have nickel-dependent enzymes in our bodies, because it is too complicated for the organism to provide the right amount."
Nevertheless, the bacteria specifically resort to the tricky nickel to initiate the hydrolysis. "Dealing with nickel is no trivial matter for the bacteria either," explains biochemist Dr Malte Sinn, "they need two auxiliary enzymes to incorporate nickel into the enzyme." The water "primed" by nickel ions in the active centre of the enzyme attacks the guanidine and converts it into ammonia and urea. The urea can in turn be converted into ammonia by further enzymes. Both compounds can thus subsequently be exploited as nitrogen sources, making the nitrogen available for building new biomolecules.
Structural images
Olga Mayans’s research team carried out structural analyses to investigate the process at the molecular level. The high specificity of the process was another surprise. The structural images show how precisely the enzyme encloses its substrate guanidine. "It has a very beautiful structure, strikingly symmetrical. The active site is very small and perfect for holding the small guanidine molecule in the correct position for hydrolysis," explains biologist Dr Jennifer Fleming from Olga Mayans’s research team.
For the research team, the current results are a first step towards understanding naturally occurring guanidine in more detail: how it is formed, what functions it has in nature, and which other organisms can utilize it. Despite its wide distribution, guanidine is still a blank spot on the biochemical map.
Key facts:
- EMBARGOED UNTIL 9 MARCH 2022, 17:00 CET (16:00 LONDON TIME, 11:00 US EASTERN TIME)
- Original publication: D. Funck, M. Sinn, J. Fleming, M. Stanoppi, J. Dietrich, R. López-Igual, O. Mayans, and J. S. Hartig: Discovery of a Ni2+-dependent guanidine hydrolase in bacteria. Nature, 2022
DOI: 10.1038/s41586-022-04490-x Link: https://www.nature.com/articles/s41586-022-04490-x - Research project of the University of Konstanz with the participation of the Instituto de Bioquímica Vegetal y Fotosíntesis, Universidad de Sevilla, and C.S.I.C, Seville.
- Funded by the ERC Consolidator Grant "RiboDisc" to Professor Jörg S. Hartig.