Fungal infection and biotrophy in rust fungi
Phytopathogenic fungi have developed a number of different strategies to attack and infect their host plants.
Fungal infection, leading to the differentiation of haustoria
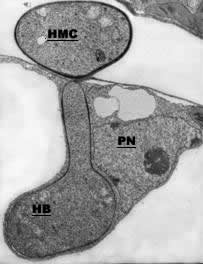
The germ tubes of rust fungi can find the stoma and produce an appressorium over the stomatal pore. A penetration hypha produced at the bottom of the appressorium pushes the guard apart and the fungus enters the substomatal cavity. After further growth, the fungus contacts the host parenchymous cell and differentiates the haustorium mother cell. From the haustorial mother cell, the rust fungus penetrates into the host cell and differentiates the haustorium with the haustorial neck and the haustorial body. For a review of haustorial structure and function see (Voegele and Mendgen, 2003; Voegele, Hahn and Mendgen, 2009 ; Voegele and Mendgen, 2011
Rust Haustoria, the center of biotrophy
Structural and functional pictures of a haustorium
 |
 |
|
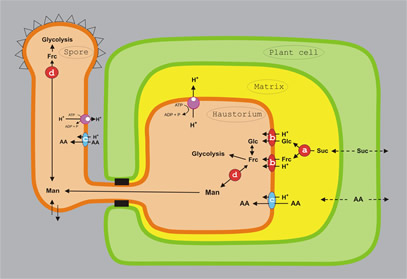
| Electron micrograph of a haustorial mother cell (HMC) and a haustorial body (HB) with neck. Note that the host plant nucleus (PN) is situated close to the haustorium | Model for amino acid and hexose uptake and redistribution in rust fungi. Schematic representation of a fungal spore, an intercellular hypha, and an haustorium (orange), an infected plant cell (green), and the extrahaustorial matrix (yellow). a) invertase INV1p; b) hexose transporter HXT1p; c) amino acid transporters AAT1p and AAT2p; d) major alcohol dehydrogenase MAD1p; Glc: D-glucose; Frc: D-fructose; Man: D-mannitol; Suc, sucrose; AA: amino acids. Solid arrows specify confirmed enzymatic conversions or transport processes, dotted arrows indicate postulated solute fluxes. |
Biosynthetic Capacities of Haustoria
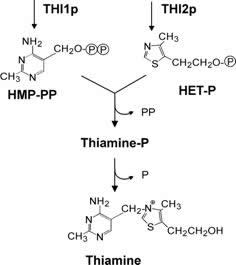
Haustoria do not only seem to be responsible for the acquisition of nutrients for the pathogen (see below), they also seem to have important biosynthetic functions. For example, a high activity in vitamin B1 biosynthesis seems to contribute to a number of pathways in sugar and amino acid metabolism.
 |
 |
|
Thiamine biosynthesis in fungi. The presumptive roles of U. fabae gene products THI1p and THI2p are indicated. HMP-PP: 2-methyl-4-amino-5-hydroxymethylpyrimidine-pyrophosphate; HET-P: 4-methyl-5-(ß-hydroxyethyl) thiazol phosphate. |
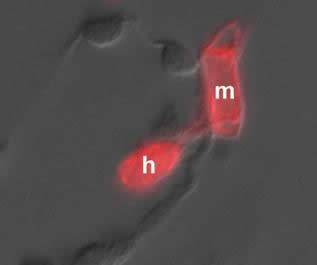
Immunolocalization of an enzyme for vitamin B1 synthesis (THI1p) in haustorial mother cell (m) and haustorium (h). |
Nutrient Uptake
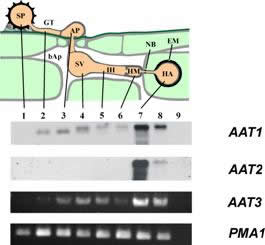
Our analysis of the capacity of U. fabae haustoria to contribute to nutrient uptake focuses on amino acids and carbohydrates. We have identified and characterized three distinct amino acid transporters (AAT1, AAT2, AAT3) and one monosaccharide transporter (HXT1).
The three amino acid transporters identified so far exhibit differential regulation of transcription, localization of the gene products, and substrate specificities. Struck et al. 2004.
A
|
 |
 |
|
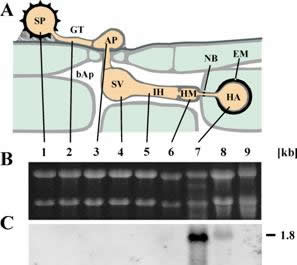
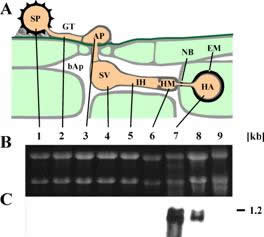
| AATx transcript distribution: A: schematic representation of rust infection structures. B: Northern Blot or RT-PCR results for the indicated genes. C. Loading control using PMA1 in RT-PCR. 1, uredospore; 2, germtube; 3, appressorium; 4, substomatal vesicle; 5, infection hyphae; 6, haustorial mother cell; 7, isolated haustoria; 8, infected leaves; 9, non-infected leaves; bAp: bulk apoplast; NB: neckband; EM: extrahaustorial matrix. The number on the right gives the size estimate in kb. |
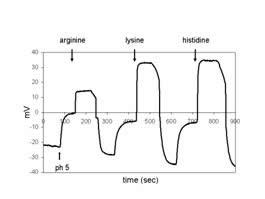
Voltage trace of AAT1p analysis in Xenopus oocytes: The addition of the respective amino acids is indicated. |
||
 |
 |
|
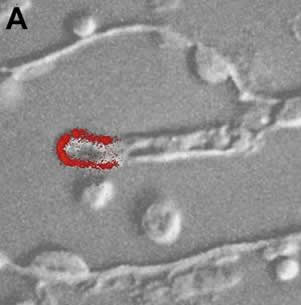
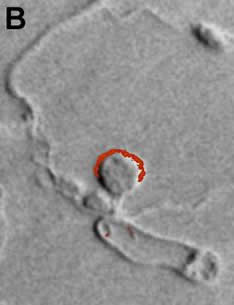
Distribution of AAT2p in monokaryotic (A) and dikaryotic (B) rust haustoria. |
||
For the monosaccharide transporter, HXT1, we were able to show that transcript as well as gene product are exclusively localized in haustoria.
 |
 |
|
| HXT1 transcripts are only found in haustoria and infected leaves. A: schematic representation of rust infection structures. B: loading control. C: Northern Blot. 1, uredospore; 2, germtube; 3, appressorium; 4, substomatal vesicle; 5, infection hyphae; 6, haustorial mother cell; 7, isolated haustoria; 8, infected leaves; 9, non-infected leaves; bAp: bulk apoplast; NB: neckband; EM: extrahaustorial matrix. The number on the right gives the size estimate in kb. |
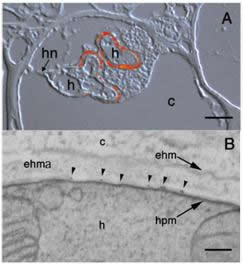
Localization of HXT1p in the periphery of fully developed haustoria (A) and along the haustorial plasma membrane (B) using S651p. A: Superimposed interference contrast and fluorescence images. h: haustorium; hn: haustorial neck; c: plant cell. Bar = 5 µm. B: Electron micrograph depicting gold labeling along the haustorial plasma membrane (hpm), but no labeling over the haustorium (h), extra haustorial matrix (ehma), extrahaustorial membrane (ehm), or plant cell (c). Bar: 0.1 µm. |
These experiments together with a detailed biochemical characterization of the transporter provided the first evidence that rust haustoria are indeed carbohydrate uptake devices.
Hexose Mobilization
Another aspect of our work is concerned with the mobilization of carbohydrates. We have good evidence for the participation of plant and fungal invertases in the enzymatic breakdown of sucrose. There are numerous reports of increased invertase activity upon wounding or pathogen infection in plants. However, no localization studies exist nor were there attempts to dissect the contributions of both organisms to the overall invertase activity. In the course of an EST sequencing project we have identified an Uromyces fabae invertase. At present we are working on a molecular and biochemical description of gene and gene product. We are also studying the distribution of plant and fungal invertases within infected plants.
Hexose Utilization
The metabolism of hexoses once they are taken up by the rust fungus is another research focus in our group. Again from the EST sequencing project there is good evidence that glycolysis as well as pentose phosphate cycle are operational in haustoria. Because of the regulatory roles carbohydrates can exert, the specificities of the enzymes downstream of the hexose transporter are important.
One of the enzymes we put our focus on is hexokinase/glucokinase, the entry enzyme for glycolysis. We want to find out what the specificity of the enzyme is and where it is located.
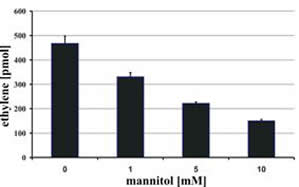
Another enzyme we scrutinize is a mannitol 2-dehydrogenase, MAD1p. This enzyme inter-converts mannitol and fructose in an NADP(H)-dependent, reversible fashion. We were able to show that MAD1p is involved in carbohydrate storage and in the quenching of reactive oxygen species generated as part of the host defense response.
 |
 |
|
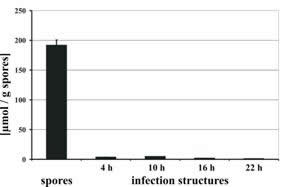
| Disappearance of mannitol from the fungal mycelium upon germination of the uredospores. | Quenching of ROS through increasing concentrations of mannitol. |
Whereas MAD1 transcripts were only localized in haustoria, MAD1p was found in the lumen of haustoria and the lumen of spores.
 |
 |
|
| MAD1 transcripts are only found in haustoria and infected leaves. A: schematic representation of rust infection structures. B: loading control. C: Northern Blot. The number on the right gives the size estimate in kb. |
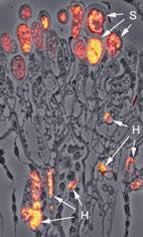
Localization of MAD1p in the lumen of haustoria and spores, using S717p. Superimposed phase contrast and fluorescence image. H: haustorium; S: spores (magnification 650-fold). |
Our results classify MAD1p as an important multipurpose enzyme in U. fabae.
Proteins from the Fungus Plant Interface
The interface between haustoria and the infected host cell represents an ideal trading post for the exchange of nutrients and information. We therefore initiated the entirety of proteins secreted from a haustorium. Our goal is to identify and characterize novel proteins, which might be linked to biotrophy or at least to the pathogenicity of rust fungi. Proteins for example like RTP1p, see below. Link and Voegele, 2008.
PTP1p, a Fungal Protein Transferred to the Host Cell
In the course of screening proteins, which were predicted to be secreted, we identified a protein, designate RTP1p. RTP1p showed a surprising localization when probed with different antibodies. Immunofluorescence data suggest that RTP1p is transferred from the extrahaustorial matrix to the cytoplasm and the nucleus of the infected host cell. This would be the first example for a fungal effector protein to be transferred to the host. Currently we are characterizing RTP1p on a biochemical basis and are trying to establish the mode of delivery. Kemen et al., 2005.
Contact: Ralf Voegele
 |